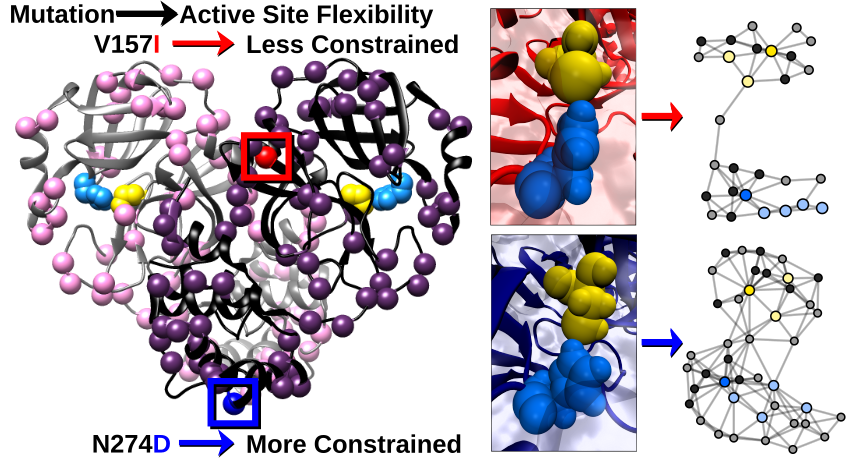
The main protease (Mpro) of SARS-CoV-2, the virus behind the global Covid-19 pandemic, is an essential part of the virus’ reproduction cycle once it has infected human cells. After viral RNA is transcribed into a long poly-protein, Mpro frees itself and other viral proteins from the poly-protein. NCASD researchers Liz Diessner and Carter Butts, in collaboration with the Martin Lab, studied the structure and dynamics of Mpro and its active site using MD simulations as well as social network methods. Metrics such as k-core and degree provide information about the cohesion of the structure, or how tightly packed the residues of the protein are to one another. When calculated over a MD simulated trajectory, these metrics give insight to the dynamics of the protein and its active site. Analysis using social network methods was done for 79 variants of the Mpro sequence, providing a glimpse into how the protein adapts to fit the environment of the human cell at the beginning of the pandemic.