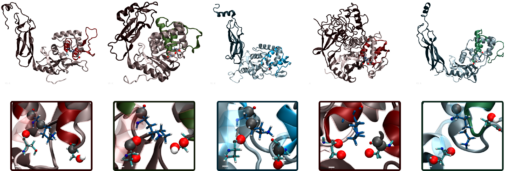
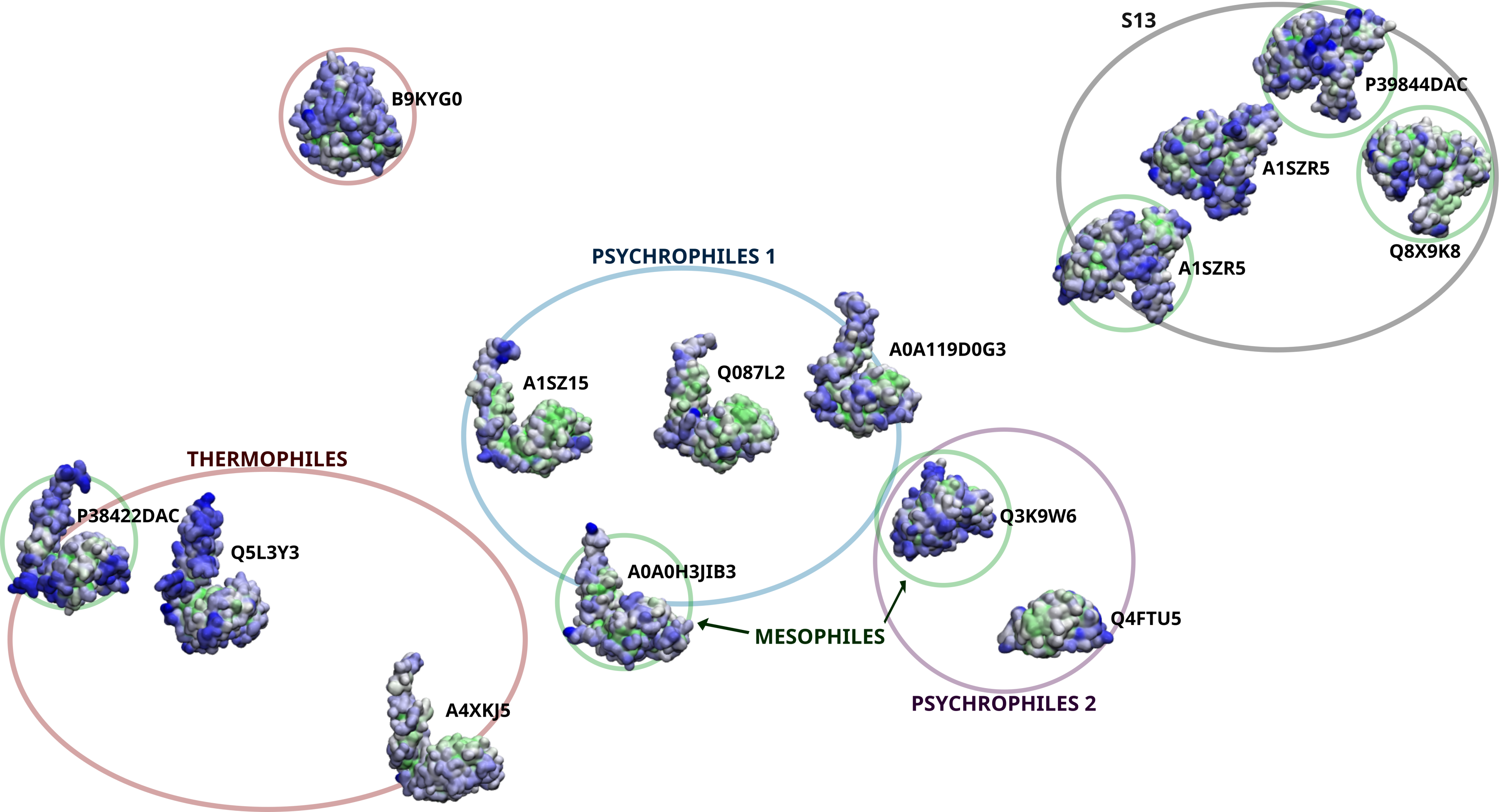
Extremophiles are organisms that thrive, or at least tolerate, extreme environmental conditions, such as the high temps and pressures found near under-water hydrothermal vents, or the freezing temperatures of permafrost soils. To survive these uncomfortable climates, organisms have had to undergo adaptations, specifically molecular adaptations that change the behavior between their biomacromolecules to reflect the thermodynamic changes that accompany extreme environments. Previously, adaptations were assumed to follow certain trends, such as increasing the number of hydrophobic residues at higher temperatures to take advantage of the 'hydrophobic effect' to maintain structure. But nature does not always follow such intuitive trends, as we show in the case of D-ala-D-ala-carboxypeptidases, an enzyme that is involved in building the cell walls of bacteria. In their recent paper, published in Biomolecules, NCASD member Elizabeth M. Diessner and colleagues use protein structure prediction, molecular modeling and dynamics simulations, and machine learning techniques to probe the differences in structure and dynamics of this set of bacterial serine proteases from a large range of extreme environments, and find trends that contradict some of the beliefs that are still being used to guide experimental research.